ASpedia2: splicing gene signatures to modulate pathway
extracted by text-mining and machine-learning
extracted by text-mining and machine-learning
ASpedia is a comprehensive database for human alternative splicing to encompass functional sequences on splicing regions and splicing gene signatures to modulate pathways.
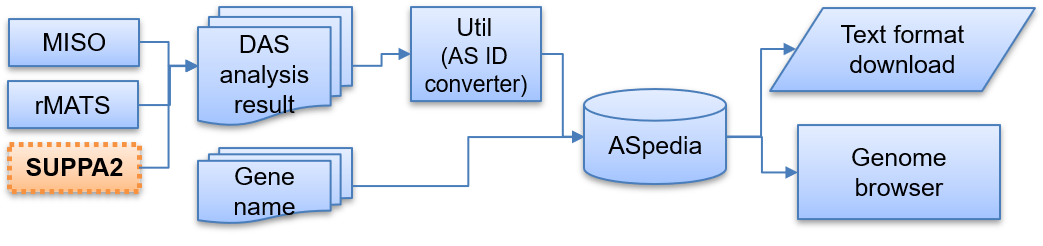
The previous version provided multi-omics profiles on splicing regions for reference genome hg18 and 19. The retrieval system allowed the search from multiple AS event IDs to annotate AS events. Multiple AS events of differential AS analysis (MISO and rMATS) could be annotated using our system.
In ASpedia2, we updated previous systems and established new databases. (1) The previous version is now expanded to human genome GRCh38, and (2) retrieval utils additionally support the SUPPA2 ID system. (3) Genome browser is also revised to UCSC genome browser to improve user convenience. AS signature databases to delineate pathways are established. (4) Knowledge-based AS signature database is established from literature using text-mining technology. (5) AS signatures to modulate cancer pathways are identified from pan-cancer transcriptome using machine-learning models. In order to develop reliable database collection, splicing signatures were assessed by cross-evaluation between transcriptome- and knowledge-based collections, predictive performance evaluation, and comparison with external differential AS analysis results.
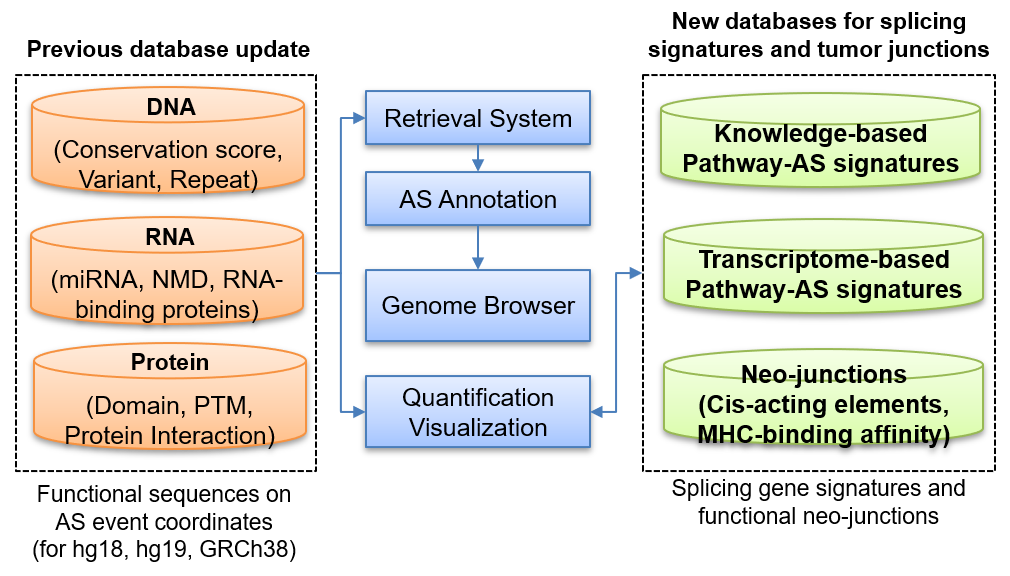
ASpedia2 supports an updated retrieval system for multiple AS events like previous version and is useful to annotate genomic profile of AS events identified by differential AS (DAS) analysis. The AS event ID system for each tool can be converted by our utility. All search results are downloadable to a text format file, and each AS event’s genomic region and functional sequence profile can be checked through the genome browser. Current update expanded our database to GRCh38 human genome reference to increase data domain coverage.
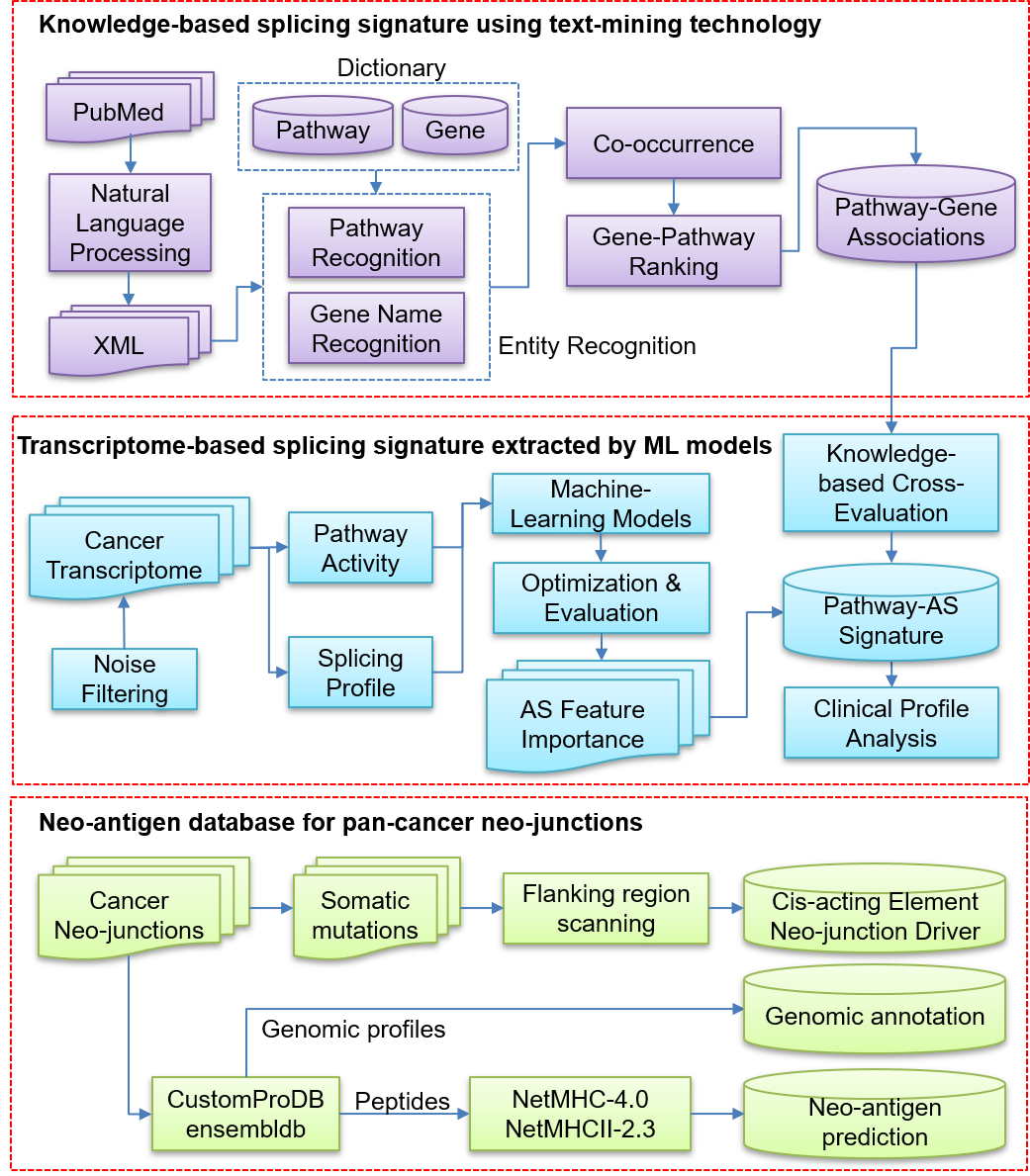
We introduce new contents of knowledge- and transcriptome-based splicing signatures and cancer neo-junctions. (1) Knowledge-based splicing signatures were extracted from literature using text-mining technology. Each signature set was collected, showing co-occurrence with the pathway term. The importance of pathway-gene pairs was ranked by multiple co-occurrence tests. (2) Transcriptome-based splicing signatures were extracted from pan-cancer transcriptome profiles using machine-learning technology. Especially, transcriptome-based signatures passed cross-evaluation from knowledge-based signatures and considered AS event recurrence detected across cancer types. (3) Cancer neo-junctions derived from variants on cis-acting elements play a critical role as a neo-antigen to bind with major histocompatibility complex (MHC). We acquired neo-junction coordinates from a previous study of the pan-cancer transcriptome [PMID: 30078747]. Genomic profiles of neo-junctions were annotated, encompassing gene name, ID, and protein-coding frame. The binding affinities between MHC class I/II and translated peptides for each neo-junction were predicted. It could be useful to explore the functionally important neo-junctions for cancer cells.